Injection frames
Minke is able to create frame files which contain injections, which can then be used to do things like characterising parameter estimation algorithms.
It is possible to do this by writing a script.
However, Minke also provides a function to do this, and a commandline interface.
It is also possible to produce injection frames as a step in an asimov workflow.
Script
from minke.noise import AdvancedLIGO
import astropy.units as u
from minke.models.cbc import IMRPhenomPv2
from minke.detector import AdvancedLIGOHanford
noise = AdvancedLIGO()
noise_ts = noise.time_series(duration=4, sample_rate=16384, epoch=998)
model = IMRPhenomPv2()
parameters = {"mass_ratio": 0.7, "total_mass": 100*u.solMass, "luminosity_distance": 100*u.megaparsec, "gpstime": 1000}
data = model.time_domain(parameters, times=noise_ts.times)
f = data['plus'].plot()
f.savefig("waveform.png")
detector = AdvancedLIGOHanford()
projected = data.project(detector,
ra=1, dec=0.5,
iota=0.4,
phi_0=0,
psi=0
)
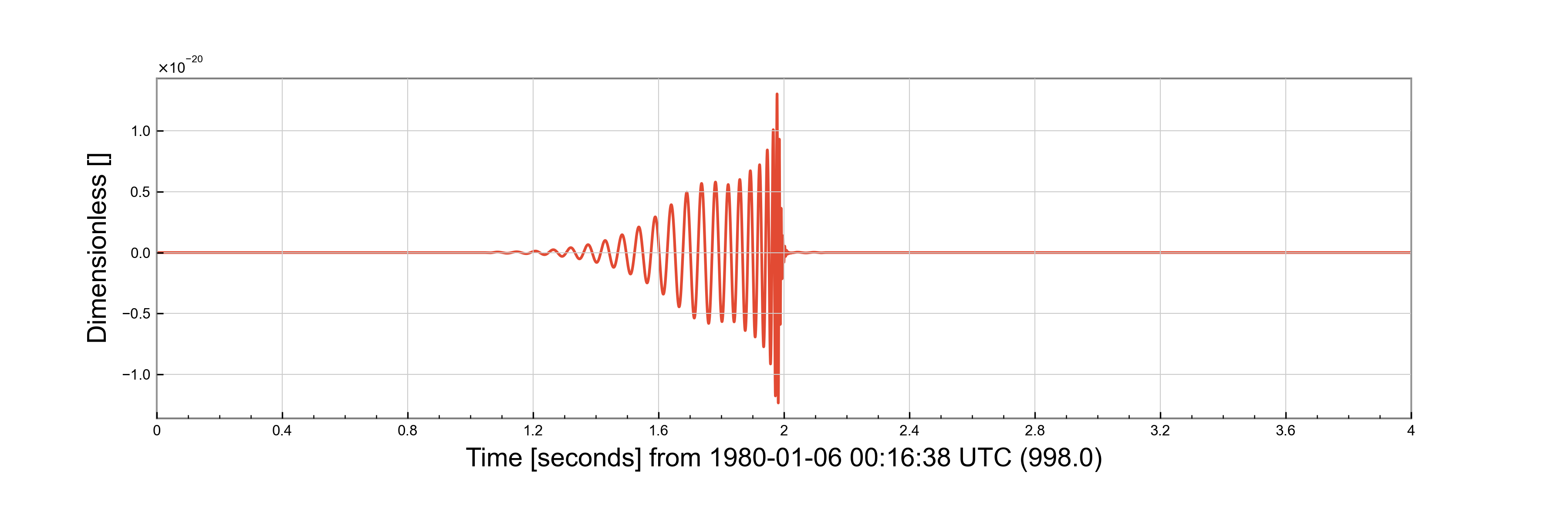
injection = (noise_ts + projected)
f = injection.plot()
f.savefig("projected_injection.png")
injection.write("test_injection.gwf", format="gwf")
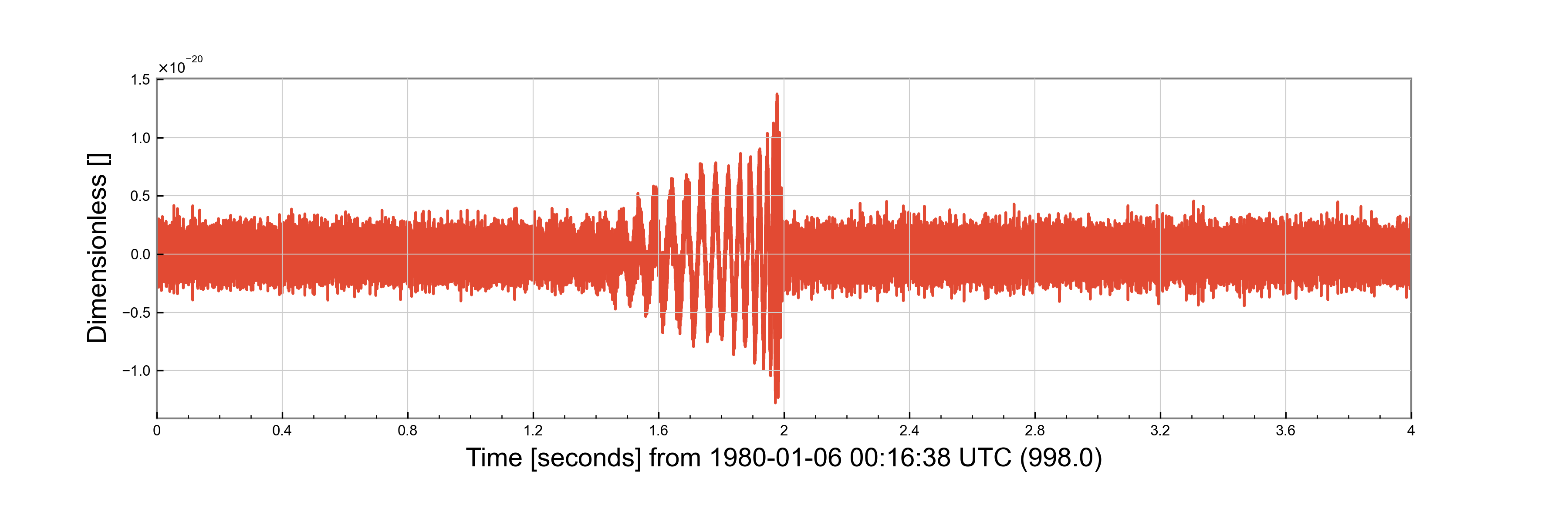
The make_injections Functions
from minke.injection import make_injection
from minke.noise import AdvancedLIGO
from minke.models.cbc import IMRPhenomPv2
from minke.detector import AdvancedLIGOHanford
import astropy.units as u
detectors = {"AdvancedLIGOHanford": "AdvancedLIGO"}
parameters = {"mass_ratio": 0.7,
"total_mass": 100*u.solMass,
"luminosity_distance": 100*u.megaparsec,
"ra": 1,
"dec": 0.5,
"iota": 0.4,
"phi_0": 0,
"psi": 0,
"gpstime": 1000}
injection = make_injection(detectors=detectors, injection_parameters=parameters, duration=4, sample_rate=4096, epoch=998)['H1']
f = injection.plot()
f.savefig("injection_script.png")
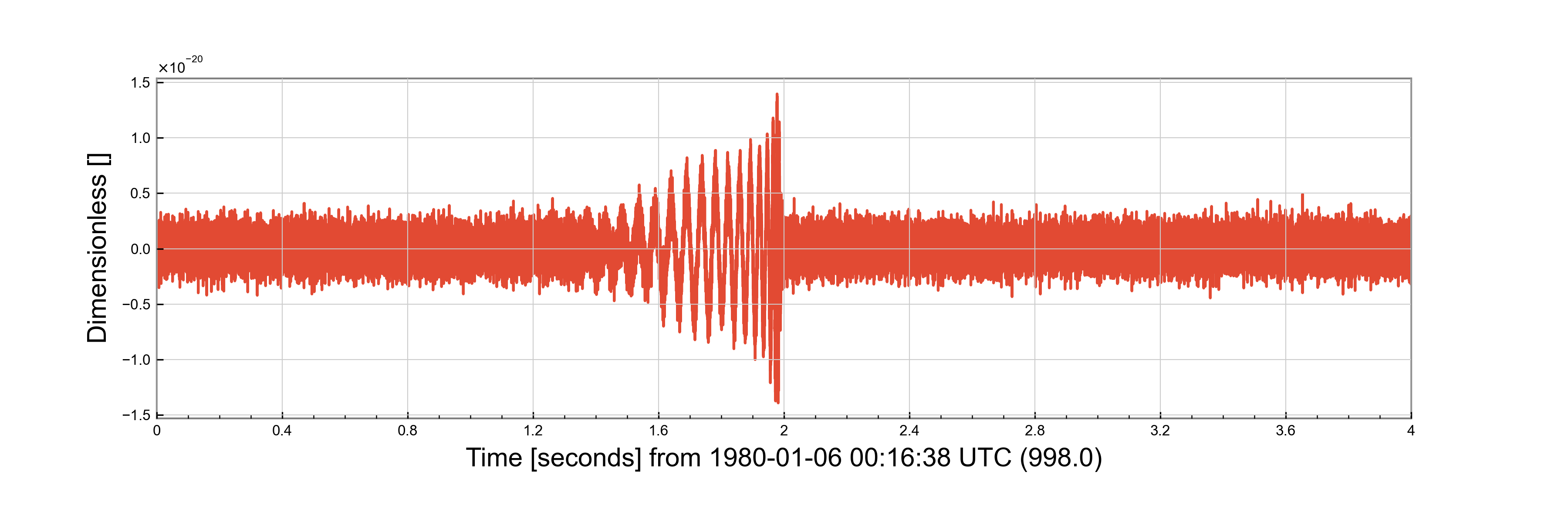
We can also produce injections into zero noise:
from minke.injection import make_injection_zero_noise
from minke.noise import AdvancedLIGO
from minke.models.cbc import IMRPhenomPv2
from minke.detector import AdvancedLIGOHanford
import astropy.units as u
detectors = {"AdvancedLIGOHanford": "AdvancedLIGO"}
parameters = {"mass_ratio": 0.7,
"total_mass": 100*u.solMass,
"luminosity_distance": 100*u.megaparsec,
"ra": 1,
"dec": 0.5,
"iota": 0.4,
"phi_0": 0,
"psi": 0,
"gpstime": 1000}
injection = make_injection_zero_noise(detectors=detectors, injection_parameters=parameters, duration=4, sample_rate=16384, epoch=998)['H1']
f = injection.plot()
f.savefig("injection_function_zero.png")
The Command-line interface
To use the command line interface you’ll need to create a YAML-formatted configuration file.
For example
injection:
duration: 4
sample_rate: 4096
epoch: 998
channel: Injection
parameters:
luminosity_distance: 400
m1: 35
m2: 30
waveform: IMRPhenomPv2
interferometers:
H1: AdvancedLIGOHanford
L1: AdvancedLIGOLivingston
psds:
H1: AdvancedLIGO
L1: AdvancedLIGO
Save this file as settings.yml.
You can then create the injection files by running minke injection --settings settings.yml
As a step in an asimov workflow